Vector Search
Rockset is a vector database. Utilize vector search to help run your RAG, semantic search, and recommendation use cases. Using vector search on Rockset allows you to seamlessly mix similarity searches over your embeddings with complex joins, selective predicates, and everything else you expect from SQL on Rockset. (Learn more: FAQ - What is Vector Search?)
Check out this blog post on 5 early adoptions of Vector Search to learn more about use cases and implementation considerations.
Looking for examples written in Python? Check out our detailed recipes that implement Vector Search for real-world use cases:
Generate embeddings
In order to execute a vector search, you will need to convert your textual data into machine-readable numerical vectors - known as embeddings. (Learn more: FAQ - What are embeddings?)
Large Language Models (LLMs) including Hugging Face and OpenAI have made embedding algorithms easily accessible and affordable. Such third party language models typically have simple to use integrations with Python orchestration tools like LangChain. (Learn more: FAQ - What resources can I use to embed my data?)
Check out our LangChain documentation for a tutorial on how to use LangChain to embed your Rockset data using our Rockset LangChain integration.
Store Embeddings in Rockset
The next step is to ingest the embeddings created in the previous step into a Rockset Collection. In the Ingest Transformation, be sure to add a VECTOR_ENFORCE function over your embeddings field. This function will ensure that all incoming vectors are uniform in length and type and will return a NULL value on failure. In addition to performing uniformity checks, it signals to Rockset that this array should be treated as a vector allowing Rockset to make indexing performance optimizations.
In the example below, we are applying VECTOR_ENFORCE over the book_embeddings field ensuring each vector is of length 1536 and type ‘float’:
SELECT
title,
author,
VECTOR_ENFORCE(book_embedding, 1536, 'float') as book_embedding
FROM
_input
Console Instructions
In the Collections tab of the console, select “Create Collection” and follow the prompts to create a collection from a File Upload, one of our fully-managed integrations, or an empty collection that you can write data to using the WriteAPI. In the “Transform Data” step, you will be prompted for an Ingest Transformation where you will need to add the VECTOR_ENFORCE function for your embedding field (refer to the example above). Continue following the rest of the Create Collection flow prompts to finish creating the collection to store your embeddings.
API Instructions
Use the Create Collection API endpoint, to create a collection in Rockset. The field_mapping_query is the Ingest Transformation query applied to the ingesting data. In this query, be sure to add the VECTOR_ENFORCE function for your embedding field.
"field_mapping_query": {
"sql": "SELECT title, author, VECTOR_ENFORCE(book_embedding, 1536, 'float') as book_embedding FROM _input"
}
KNN Search
At this point, we can perform a K-Nearest Neighbors (KNN) search. KNN search, also referred to as exact search, is a linear search that involves computing the distance/similarity between a query vector and all other vectors and selecting the k nearest neighbors. A simple KNN search query can be written using an ORDER BY clause over a distance/similarity function and a LIMIT over how many nearest neighbors to select. Rockset supports 3 distance functions for k-NN search: EUCLIDEAN_DIST, COSINE_SIM, and DOT_PRODUCT. (Learn more: FAQ - What is KNN?)
In the example below, we are retrieving the top 10 books (k=10 nearest neighbors) with the smallest cosine similarity between our target_embedding parameter and our book_embedding field. The parameter value will be an embedding of a title or text you are searching for. Be sure to use the same embedding model that you used to embed the original data.
SELECT
title,
author
FROM
book_dataset
ORDER BY
COSINE_SIM(:target_embedding, book_embedding) DESC
LIMIT
10
Console Instructions
In the Query Editor tab of the Console, write a query similar to the above. To create a :target_embedding parameter, click the + button following “Parameters” and populate the fields as shown in the screenshot below. Hint “Enter” on your keyboard or click away from the “Add Parameter” modal to save the parameter. Click “Run” to execute your KNN search query.
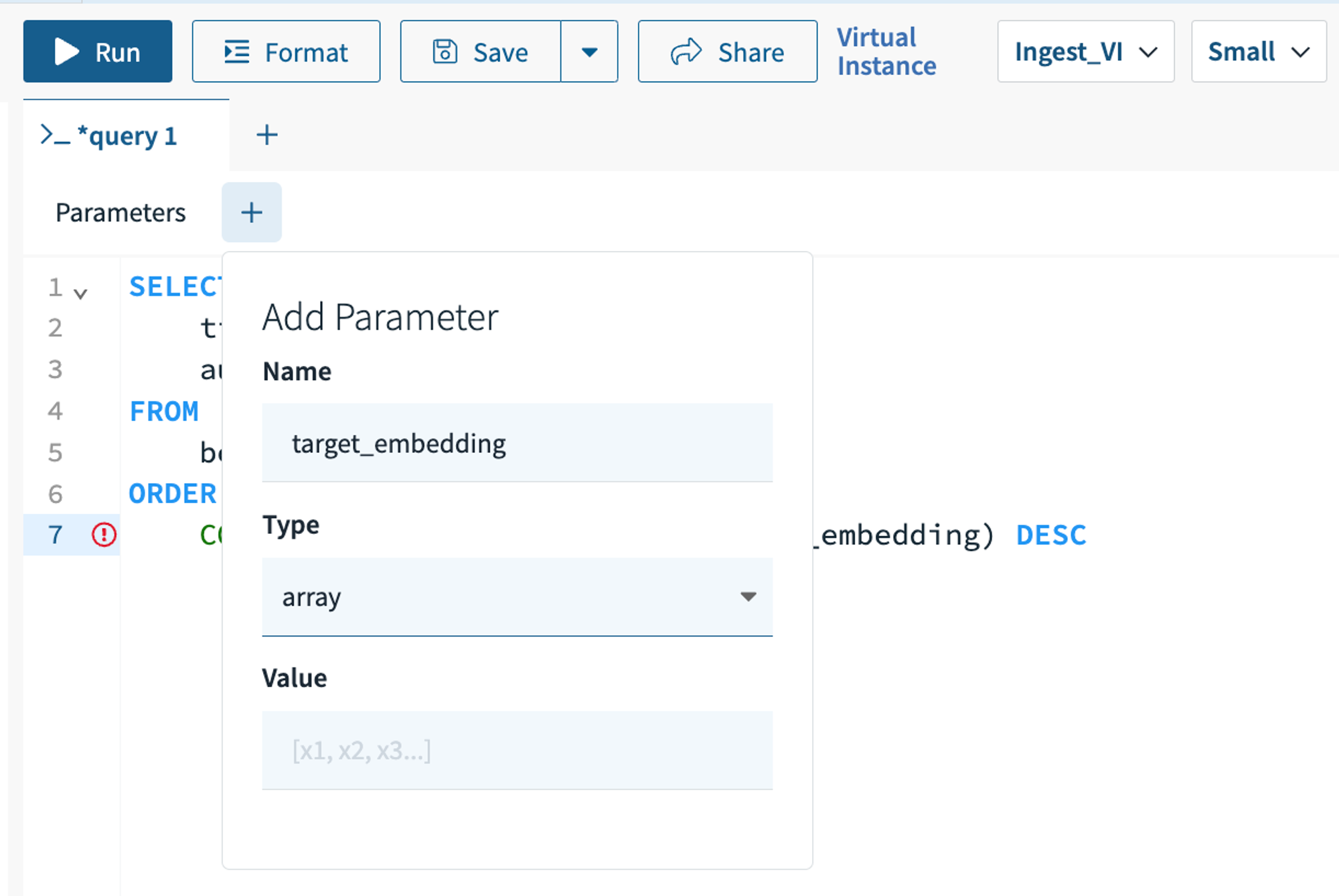
API Instructions
Use the Execute SQL Query API endpoint, to execute a query similar to the above. In the sql object, query will be a string containing the SQL query and parameters is an array of parameter objects. You will need to pass one parameter for the target_embedding with the following format:
"parameters": [
{
"name": "target_embedding",
"type": "array",
"value": "<YOUR_SEARCH_EMBEDDING>"
}
]
Similarity Index
A major downside of using KNN search is that its computationally intensive. In many use cases, we can approximate the results and utilize another algorithm called Approximate Nearest Neighbor (ANN) search. In order to implement ANN search, we will need to build an additional index: the Similarity Index. (Learn more: FAQ - What is ANN?)
Creating the index
To create the Similarity Index, we can run the following DDL command:
CREATE
<SIMILARITY|DISTANCE> INDEX <NAME>
ON
FIELD <Collection|RRN>:<FieldPath> DIMENSION <Vector Dimension> AS <Factory String>
<SIMILARITY|DISTANCE>: Which metric to order the vectors by. This metric must be decided at creation time and will affect what functions can utilize the index.<NAME>: Name for the new index.<Collection|RRN>: Either the collection name or the collection RRN of the collection we wish to build an index for.<Vector Dimension>: The dimension of the vectors in said collection.<Factory String>: Specifies underlying type information and configuration for the new index. Has the form:<Provider>:<Parameters>:<ProviderConfig><Provider>: The library or creator of the index implementation.<Parameters>: Rockset-specific parameters for querying and maintaining the index. Assigning a parameter value has the syntaxparam=value. Setting parameters is optional.- Available parameters include:
- nprobe: The number of centroids (posting lists) that will be visited during a query by default (1 by default). (Learn more: FAQ - How do you specify a centroid value?)
- minprobe, maxprobe: Specify a minimum number of posting lists to traverse and expand the search as necessary to fulfill a passed in limit up until maxprobe lists have been traversed. *Not yet available.
- Available parameters include:
<ProviderConfig>: Provider-specific index construction string.- Construction strings for providers:
- faiss: This string is defined by the FAISS library for index factory construction. A formal outline of the grammar for the factory string can be found here. FAISS supports a few form of indexes but Rockset only supports the IVF family of indexes mentioned here.
- Construction strings for providers:
In the example below, we are creating a Similarity Index for our book embeddings using the FAISS IVF index with 256 centroids and a Flat coarse quantizer:
CREATE
SIMILARITY INDEX book_catalogue_embeddings_ann_index
ON
FIELD commons.book_dataset:book_embedding DIMENSION 1536 AS 'faiss::IVF256,Flat';
Note: we did not specify any parameters so the default nprobe value for the index will be 1.
Similarity indexes are built using the Ingest VI.
DDL Command in Console Instructions
In the Query Editor tab of the Rockset Console, write a similar DDL Command to the above. Click “Run” to execute the query.
Create via Collections Tab in Console Instructions
In the Collections tab of the Console, select the collection you would like to create a similarity index for. Under the “Indexes” heading, click “Create a Similarity Index”. Follow the prompts to create your desired index.

If using the example above, you would input the following parameters:
-
Name: book_catalogue_embeddings_ann_index
-
Embedding Field: book_embedding
-
Dimensions: 1536
-
Parameters: Standard, IVF, 256 centroids
-
Distance Function:
INNER_PRODUCT
API Instructions
Use the Execute SQL Query API endpoint, to execute a query similar to the above DDL command. In the sql object, query will be a string containing the DDL command.
Querying the Index
The index will take some time to be built, and we can query the status of the index to determine when we can start utilizing the index for ANN search. The CREATE DDL command in the previous step will return the index RRN, Rockset's globally unique identifier. To check the status of the index, query the _system workspace like below:
SELECT
*
FROM
_system.similarity_index
WHERE
rrn = <RRN>
This will return metadata for the index including its index_status which can be in state 'TRAINING' or 'READY'. The index is only useable once it has reached the 'READY' state.

Given the number of centroids C, Rockset must read at least C * collection shard count vectors before training completes. If not enough vectors are available to be trained on, the index will stay in the 'TRAINING' state and wait until more vectors are ingested. (Learn more: FAQ - How do you specify a centroid value?)
Index Building Tip
Building an index is a memory intensive operation as clustering is performed in memory. For large collections it is recommended that only one similarity index be trained at a time.
Once the command finishes, vector search queries will be able to transparently take advantage of the new index.
Console Instructions
In the Query Editor tab of the Rockset Console, write a similar SQL query to the above. Click “Run” to execute the query.
API Instructions
Use the Execute SQL Query API endpoint, to execute a query similar to the above. In the sql object, query will be a string containing the SQL query.
Deleting the index
To delete an index, use the DROP command. Once performed, queries will no longer use the index and Rockset will begin the process of cleaning up the index. Please note that deleting an index will inquire some CPU load.
DROP <SIMILARITY|DISTANCE> INDEX <Index Name>
For example, to clear the index "book_catalogue_embeddings_ann_index" that we created we would run the following query:
DROP SIMILARITY INDEX book_catalogue_embeddings_ann_index
DDL Command in Console Instructions
In the Query Editor tab of the Rockset Console, write a similar DDL Command to the above. Click “Run” to execute the query.
Delete via Collections Tab in Console Instructions
In the Collections tab of the Console, select the collection you created the similarity index for. Under the “Indexes” heading, hover over the row for the similarity index you would like to delete. A red trash icon will appear on the far right. Click this icon and complete the prompt to delete the index.
API Instructions
Use the Execute SQL Query API endpoint, to execute a query similar to the above DDL command. In the sql object, query will be a string containing the DDL command.
ANN Search
Similar to KNN search, we can execute an ANN search simply by applying an ORDER BY clause over an approximate distance/similarity function. Rockset supports 3 distance functions for k-NN search: APPROX_EUCLIDEAN_DIST and APPROX_DOT_PRODUCT. To change the number of posting lists iterated in an ANN search you may update the nprobe parameter per query by setting the option options(nprobe=<# of posting lists>). You cannot specify more posting lists to query than there are centroids in the index. (Learn more: FAQ - What is ANN?)
In the example below, we are retrieving the approximate top 30 books with the smallest dot product between our target_embedding parameter and our book_embedding field:
SELECT
book_dataset.title,
book_dataset.author
FROM
book_dataset ds
JOIN book_metadata m ON ds.isbn = m.isbn
WHERE
m.publish_date > DATE(2010, 12, 26)
and m.rating >= 4
and m.price < 50
ORDER BY
APPROX_DOT_PRODUCT(:target_embedding, ds.book_embedding) option(nprobe=2) DESC
LIMIT
30
Rockset's Cost Based Optimizer (CBO) will determine which index to use. If there is no index on the field for the corresponding approximate distance function, then it will invoke a KNN search which does not use a Similarity index.
Force Index Access with HINTs
There are situations where the CBO may have not collected enough stats to feel confident in using the Similarity index, so to force the index to be used you may add
HINT(access_path=index_similarity_search)after theFROMclause.If the index is not yet ready, approximate similarity search queries will return an error. If you would still like to query without an index you may force the optimizer to perform a brute force search using the column index with
HINT(access_path=column_scan).If there are multiple useable indexes on the same field of the collection, the oldest available index will be used for the query by default. You can override this default behavior by using
HINT(similarity_index='<Name>').
When searching, Rockset will try to push any predicate checks into the similarity index scan itself to avoid having to "pre" or "post" filter results. When querying the similarity index a provided result limit is required for the index to be used since without a limit a full scan of the collection would be performed using the index. Rockset will try to push a limit down to the similarity index, but if it fails due to a predicate that cannot be resolved within the index, then the index will not be used and the optimizer will fall back to a brute force KNN search.
Console Instructions
In the Query Editor tab of the Console, write a query similar to the above. To create a :target_embedding parameter, click the + button following “Parameter” and populate the fields as shown in the screenshot. Click “Run” to execute your KNN search query.

API Instructions
Use the Execute SQL Query API endpoint, to execute a query similar to the above. In the sql object, query will be a string containing the SQL query and parameters is an array of parameter objects. You will need to pass one parameter for the target_embedding with the following format:
"parameters": [
{
"name": "target_embedding",
"type": "array",
"value": "<YOUR_SEARCH_EMBEDDING>"
}
]
SELECT
_id,
words,
BM25(['hello', 'world'], words) as bm25_score
FROM
data
ORDER BY bm25_score DESC
Optimizing Vector Search
An important benefit to using Rockset for vector search is that joining tables and checking complex predicates can easily coexist with a similarity search. You simply update the WHERE clause and JOIN any extra information you are interested in. In vector search parlance this is commonly referred to as metadata filtering. (Learn more: FAQ - How can I make my vector search queries faster?)
SELECT
book_dataset.title,
book_dataset.author
FROM
book_dataset ds
JOIN book_metadata m ON ds.isbn = m.isbn
WHERE
m.publish_date > DATE(2010, 12, 26)
and m.rating >= 4
and m.price < 50
ORDER BY
COSINE_SIM(:target_embedding, ds.book_embedding) DESC
LIMIT
30
Want to learn more?
Check out our KNN Search Workshop and ANN Search Workshop to learn more about utilizing Vector Search in Rockset.
Check out our blog “How Rockset Built Vector Search for Scale in the Cloud” for more information on how Vector Search was integrated into Rockset.
Vector Search FAQs
What are embeddings?
Embeddings are just arrays of numbers that provide a compact, meaningful representation of large, complex, and unstructured pieces of data. These embeddings are typically generated using ML models which have the ability to ingest complex unstructured data and map the data to a compressed vector form representation of fixed length.
Representing complex data as compact arrays has many advantages including making storage and processing incredibly efficient. Another byproduct of working with embedding representations is that they are easily comparable with each other.
What resources can I use to embed my data?
Vector Search has become increasing popular due to the accessibility and advancements in large language models. These language models include: GPT models from OpenAI, BERT by Google, LaMDA by Google, PaLM by Google, LLaMA by Meta AI. The embeddings generated by these models are high-dimensional and can be stored and indexed in Rockset for efficient vector search.
What is Vector Search?
Vector Search refers to the practice of performing a similarity search over a set of vectors or "embeddings". (Learn more: What are embeddings?)
You can think of vectors as points in an N-Dimensional "latent space" and as such you can calculate the distance/similarity between two vectors using standard techniques like finding the Euclidean distance. (Learn more: How do you calculate distance/similarity in vector space?)
These distance functions calculate the semantic similarity of the data that was used to create the vectors and we call searching for vectors that are close to a specific vector "similarity search", otherwise known as vector search.
How do you calculate distance/similarity in vector space?
How close two vectors are in vector space correlates with how closely their semantic meanings are. There are three proximity functions that are commonly used.
Euclidean Distance:
- Geometrically, it measures the “straight-line” distance between two points.
- Algebraically, it’s the square root of the sum of the squared differences between the vectors.
Cosine Similarity:
- Geometrically, it quantifies the similarity of the directions, regardless of the magnitudes.
- Algebraically, it measures the cosine of the angle between two vectors.
Dot Product:
- Geometrically, it measures how closely two vectors align, in terms of the directions they point.
- Algebraically, it is the product of the two vectors' Euclidean magnitudes and the cosine of the angle between them.
What is K-Nearest Neighbors (KNN) search?
K-Nearest Neighbors (k-NN) is a simple, yet effective, machine learning algorithm used for classification and regression tasks. It operates on the principle that similar things exist in close proximity. k-NN search is a linear search that involves computing the distance/similarity between a query vector and all other vectors and selecting the k nearest neighbors. The algorithm is non-parametric, meaning it does not make any underlying assumptions about the distribution of data, making it versatile for various kinds of data, but it can become computationally intensive as dataset size grows, owing to its need to calculate distances between data points.
(Learn more: How do you calculate distance/similarity in vector space?)
How can I make my vector search queries faster?
When the dataset is small, it is easy to scan the whole dataset to perform a KNN search, and many times all of the vectors that need to be looked at can fit in memory allowing for fast scans and comparisons. However, as the dataset increases, so does the latency.
One method to speed up vector search queries is to add selective predicates to our query (in the WHERE clause). This metadata filtering can significantly reduce the size of the dataset needed to scan over.
Another method to speed up vector search queries is to implement an approximate nearest neighbors search (ANN). In some cases it may be inescapable that we want to query over a billion or more vectors, and the cost of looking up and comparing every single one of our stored vectors is too high. At a certain point an exact KNN search becomes too expensive to scale. Thankfully in most cases exact ordering is not necessary and we really only need a best effort, or Approximate Nearest Neighbor (ANN) search.
(Learn more: What is Approximate Nearest Neighbor (ANN) search?)
What is Approximate Nearest Neighbor (ANN) search?
Approximate Nearest Neighbors (ANN) search is a computational technique used to efficiently find the approximate nearest neighbors of a point in a high-dimensional space. Unlike exact searches that meticulously compute the closest points with precise accuracy, ANN search aims for high speed and reduced computational cost by allowing for a small margin of error in the results. It employs various algorithms and data structures, such as locality-sensitive hashing, trees, or graphs, to quickly approximate the nearest neighbors without exhaustively comparing every point in the dataset. This trade-off between accuracy and efficiency makes ANN search particularly valuable in large-scale and real-time applications, such as recommendation systems, image and video retrieval, and machine learning tasks where slight inaccuracies are acceptable in exchange for significant gains in performance.
How does Rockset utilize FAISS?
Rockset's index filtering utilizes FAISS Inverted File Indexes for its ANN architecture.
Rockset will train a set of centroids on the collection's vector data with the number of these centroids being N specified in the term IVF{N} used in the factory string. Each centroid acts as a representative for a cluster of similar vectors and their associated documents. Together the centroids form an index on the vector data that can be used at query time. For each centroid Rockset will have a posting list that can be used for iteration.
At query time FAISS will gather the set of centroids (clusters/posting lists) in the index that have the smallest coarse distance from the query vector. The number of centroids that are retrieved and searched are based on the nprobe parameter whose default is set at index creation but can also be specified for each individual query. From here Rockset must simply iterate the posting lists and return the closest vectors.
When searching nprobe posting lists, the results returned may be less than the requested limit due to a selective predicate. In this case increasing the centroids to probe may increase the number of returned results.
How do you specify a centroid value?
You must specify a centroid value that gives you a sufficient number of samples in each cluster based on your total number of vectors and collection shard count, as clustering of the collection when building the index is based on the shard count (Learn more: Shard Count documentation). We recommend a value that is about 16x the square root of (# vectors/collection shard count), as long as this value is no more than (# vectors/collection shard count of the collection). If it is larger, use a smaller factor than 16. If you have a relatively small collection, consider using a centroid value around ((# vectors/40) / collection shard count).
For example, if you have 16,000 vectors in a collection with 16 shards, you may use a centroid value of 505 and specify IVF505 since 16x(square root(16000/16)) = ~505.
If you are receiving an 'Insufficient samples' error, this is an indicator that you need to adjust your centroid value.
What vector datatypes does Rockset support?
Rockset vector operations support float and int type vectors.
What are the max vector dimensions?
Vectors are syntactically identical to arrays and there is no limit on array size, so there is no limit on vector size.
Is there a limit on the number of vectors?
There is no general limit on the number of vectors that can be stored in Rockset. Rockset's disaggregated Storage Architecture allows your storage tier to scale independently of your compute needs.
Can you update vectors?
Yes, you manipulate vectors in all of the same ways you would manipulate arrays. Rockset sits on top of an LSM tree based RocksdDB storage engine, which means random mutations are fast. Updates, inserts, and deletes are immediately visible in any ANN index associated with the vector.
Why use Rockset for vector search?
Rockset is already built for low-latency complex analytics on real-time data which perfectly complements vector search use cases.
Updated 10 months ago
